testdat is a programmatic implementation similar to Google/open refine

Forget big data, small data is the real revolution
- Rufus Pollock
Data checking should be something that you do all the time, but it’s normally painful and boring.
Most of data science is really cleaning and munging messy data - most data scientists I know
testdat is a programmatic implementation similar to Google/open refine
Dates
Mssing values
Outliers
Spatial bounds
Mangled strings (inconsistent capitalization, abbreviated names, pairs of elements with low distance)
library(testdat) dat <- data.frame( date = rep(as.Date("2014-01-01"),10), num = c(1:8,999,"n/a"), name = c("NULL","naa",rep("foo",8)) ) dat
→ date num name → 1 2014-01-01 1 NULL → 2 2014-01-01 2 naa → 3 2014-01-01 3 foo → 4 2014-01-01 4 foo → 5 2014-01-01 5 foo → 6 2014-01-01 6 foo → 7 2014-01-01 7 foo → 8 2014-01-01 8 foo → 9 2014-01-01 999 foo → 10 2014-01-01 n/a foo
find_NA(dat)
→ row column value → 1 9 2 999 → 2 10 2 n/a → 3 1 3 NULL
find_NA(dat)
→ row column value → 1 9 2 999 → 2 10 2 n/a → 3 1 3 NULL
class(dat$num)
→ [1] "character"
clean_dat <- fix_NA(dat, custom_NAs = "naa") clean_dat
→ date num name → 1 2014-01-01 1→ 2 2014-01-01 2 → 3 2014-01-01 3 foo → 4 2014-01-01 4 foo → 5 2014-01-01 5 foo → 6 2014-01-01 6 foo → 7 2014-01-01 7 foo → 8 2014-01-01 8 foo → 9 2014-01-01 NA foo → 10 2014-01-01 NA foo
class(clean_dat$num)
→ [1] "numeric"
test-all.Rcontext("checking for NAs") test_that("data dont contain NAs", { data_files <- lapply(dir(pattern = "*.csv"), read.csv) na_check <- sapply(data_files, test_NA) expect_true(all(na_check)) })

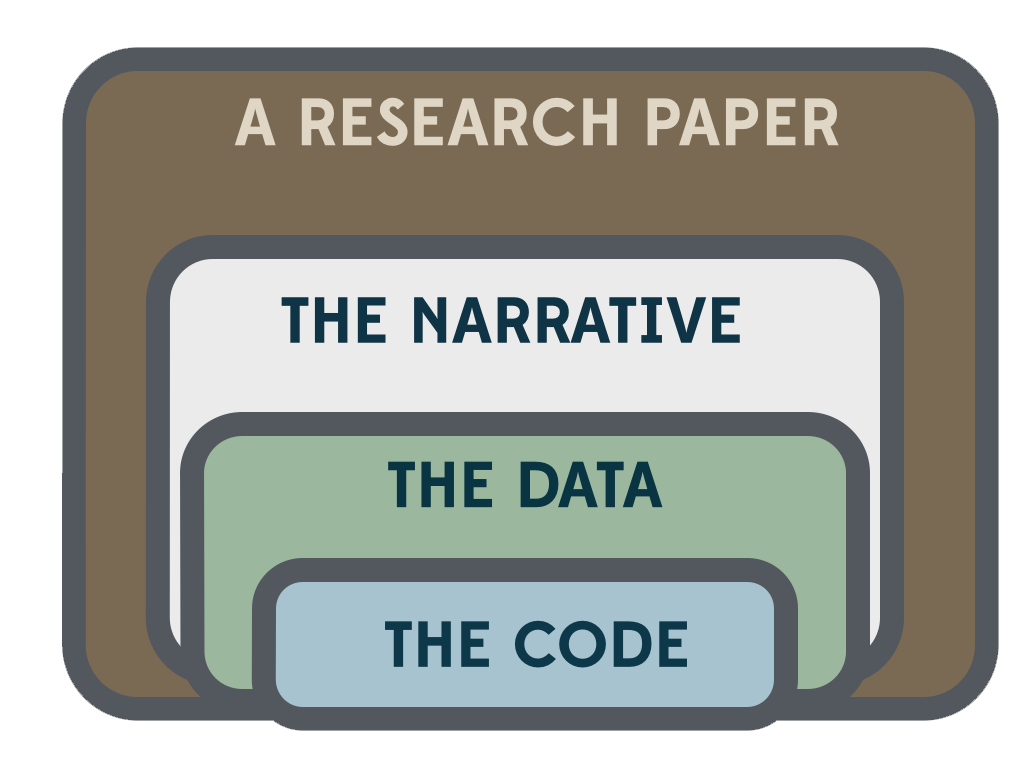
An article about [computational] science in a scientific publication is not the scholarship itself, it is merely advertising of the scholarship. The actual scholarship is the complete set of instructions [and data] which generated the figures.
- David Donoho (Stanford University)
Experimentation, theory, computing, data-intensive

Data and computer code should be made available at an early stage
 Article
Article

Enable access to scientific data repositories, full-text of articles, and science metrics and also facilitate a culture shift in the scientific community.

ropensci.org/packages
|
|
|
|


rdryadlibrary(rdryad) library(dplyr) data <- download_url("10255/dryad.1759") %>% dryad_getfile
library(rfisheries) library(dplyr) who <- c("TUX", "COD", "VET", "NPA") # Four well known commercial fisheries species_data <- function(x) of_landings(species = x) who %>% lapply(., species_data) %>% rbind_all
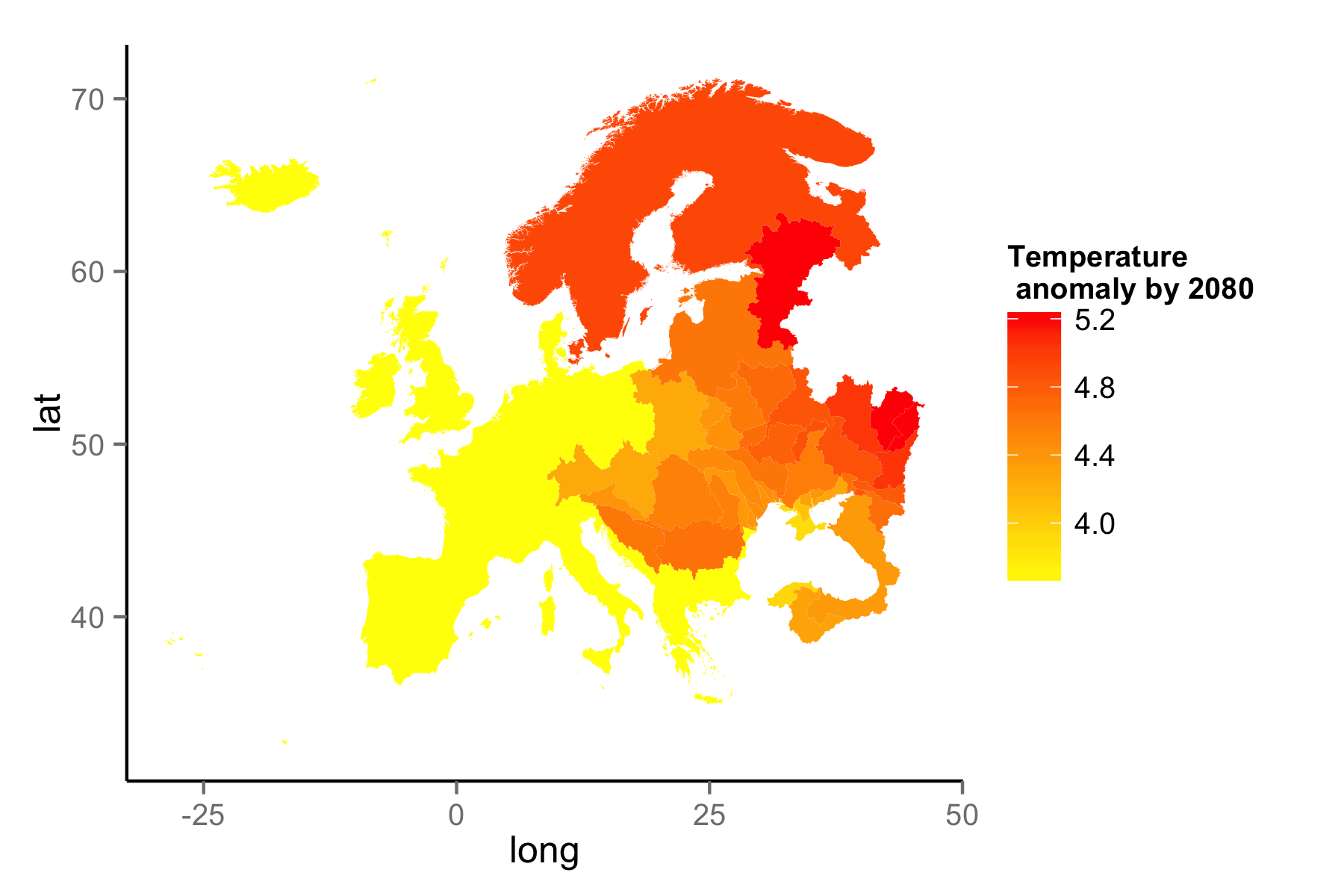
rWBclimatelibrary(rWBclimate) eu_basin <- create_map_df(Eur_basin) eu_basin_dat <- get_ensemble_temp(Eur_basin, "annualanom", 2080, 2100)

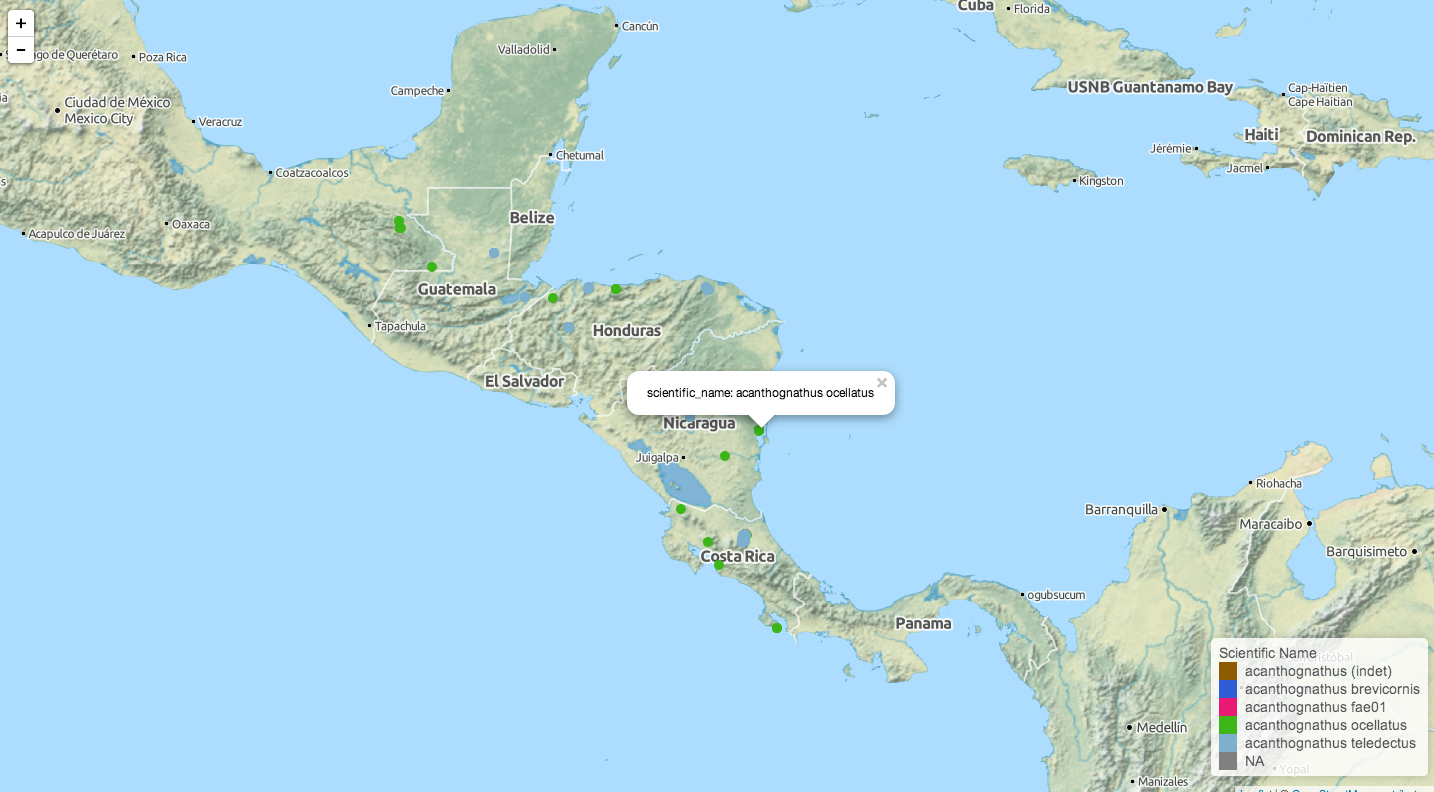
library(AntWeb); library(dplyr) aw_data(genus = "acanthognathus") %>% aw_map
plotlyplotly
(rfigshare)R and obtain a data citation.library(rfigshare) id <- fs_create("Fisheries dataset", "A dataset containing catch for 4 important commercial fish species","dataset") fs_upload(id, "dat.csv")

/