install.packages("ecoengine", dependencies = TRUE) # Requires R version 3.0.1 or higher
library(ecoengine) ee_about()
## type endpoint ## 1 meta-data http://ecoengine.berkeley.edu/api/sources/ ## 2 meta-data http://ecoengine.berkeley.edu/api/footprints/ ## 3 data http://ecoengine.berkeley.edu/api/checklists/ ## 4 data http://ecoengine.berkeley.edu/api/sensors/ ## 5 data http://ecoengine.berkeley.edu/api/vtmveg/ ## 6 data http://ecoengine.berkeley.edu/api/observations/ ## 7 data http://ecoengine.berkeley.edu/api/photos/ ## 8 actions http://ecoengine.berkeley.edu/api/search/
source_list <- ee_sources() unique(source_list$name)
## [1] "LACM Vertebrate Collection" ## [2] "MVZ Birds" ## [3] "MVZ Herp Collection" ## [4] "MVZ Mammals" ## [5] "Wieslander Vegetation Map" ## [6] "CAS Herpetology" ## [7] "Consortium of California Herbaria" ## [8] "UCMP Vertebrate Collection" ## [9] "Sensor Data Qualifiers" ## [10] "Essig Museum of Entymology"
results <- ee_observations(genus = "Lynx", progress = FALSE) results
## [Total results]: 795 ## [Args]: ## country = United States ## genus = Lynx ## georeferenced = FALSE ## page_size = 25 ## page = 1 ## [Type]: observations ## [Number of results]: 25
results <- ee_observations(genus = "Lynx", page = 1:2, progress = FALSE) results
## [Total results]: 795 ## [Args]: ## country = United States ## genus = Lynx ## georeferenced = FALSE ## page_size = 25 ## page = 1 2 ## [Type]: observations ## [Number of results]: 50
pinus_observations <- ee_observations(scientific_name = "Pinus", page = 1, quiet = TRUE, progress = FALSE) pinus_observations
## [Total results]: 48333 ## [Args]: ## country = United States ## scientific_name = Pinus ## georeferenced = FALSE ## page_size = 25 ## page = 1 ## [Type]: observations ## [Number of results]: 25
animalia <- ee_observations(kingdom = "Animalia") Artemisia <- ee_observations(scientific_name = "Artemisia douglasiana") asteraceae <- ee_observationss(family = "asteraceae") vulpes <- ee_observations(genus = "vulpes") Anas <- ee_observations(scientific_name = "Anas cyanoptera", page = "all") loons <- ee_observations(scientific_name = "Gavia immer", page = "all") plantae <- ee_observations(kingdom = "plantae") # grab first 10 pages (250 results) plantae <- ee_observations(kingdom = "plantae", page = 1:10) chordata <- ee_observations(phylum = "chordata") # Class is clss since the former is a reserved keyword in SQL. aves <- ee_observations(clss = "aves")
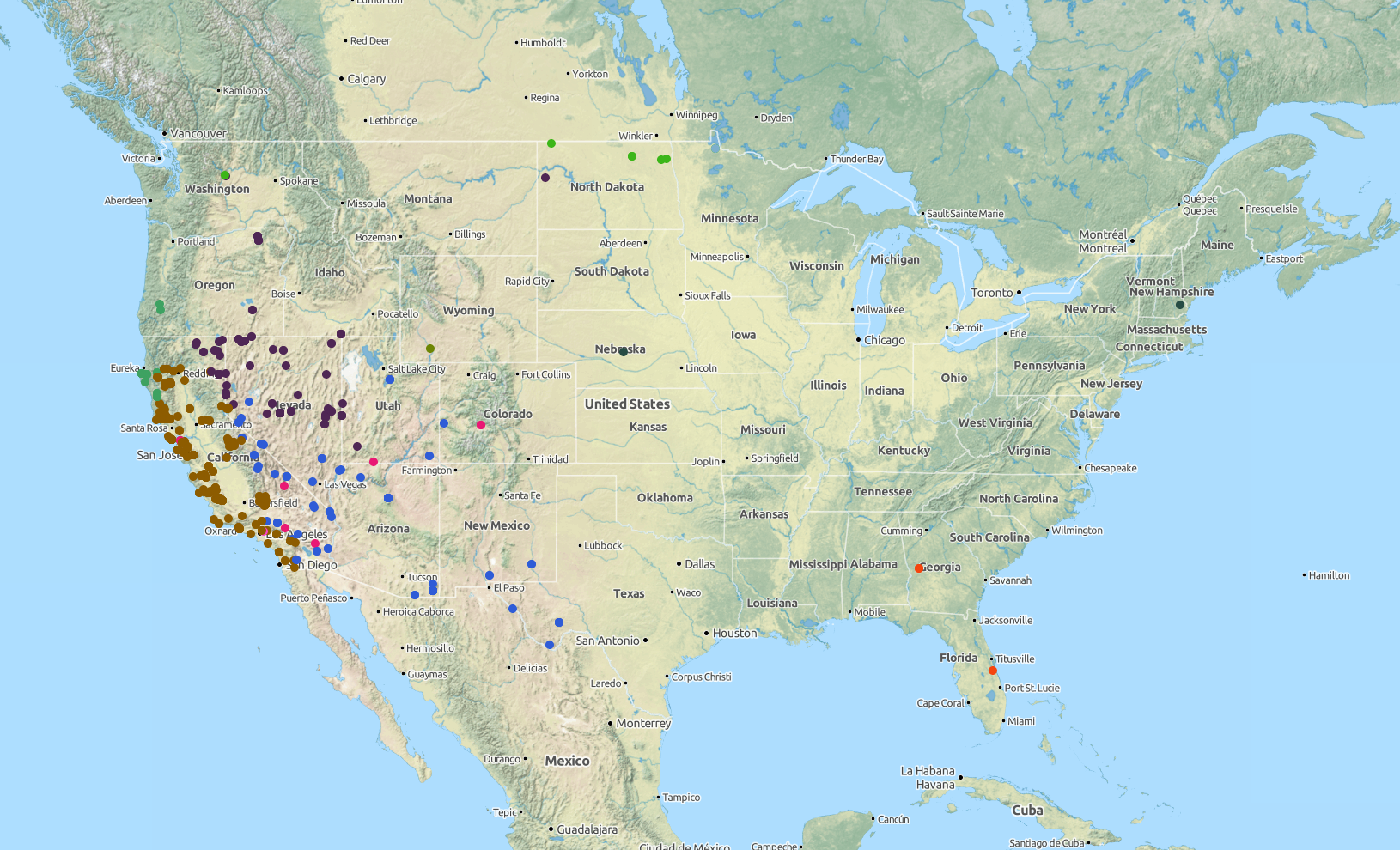
lynx_data <- ee_observations(genus = "Lynx", georeferenced = TRUE, page = "all", quiet = TRUE) ee_map(lynx_data)
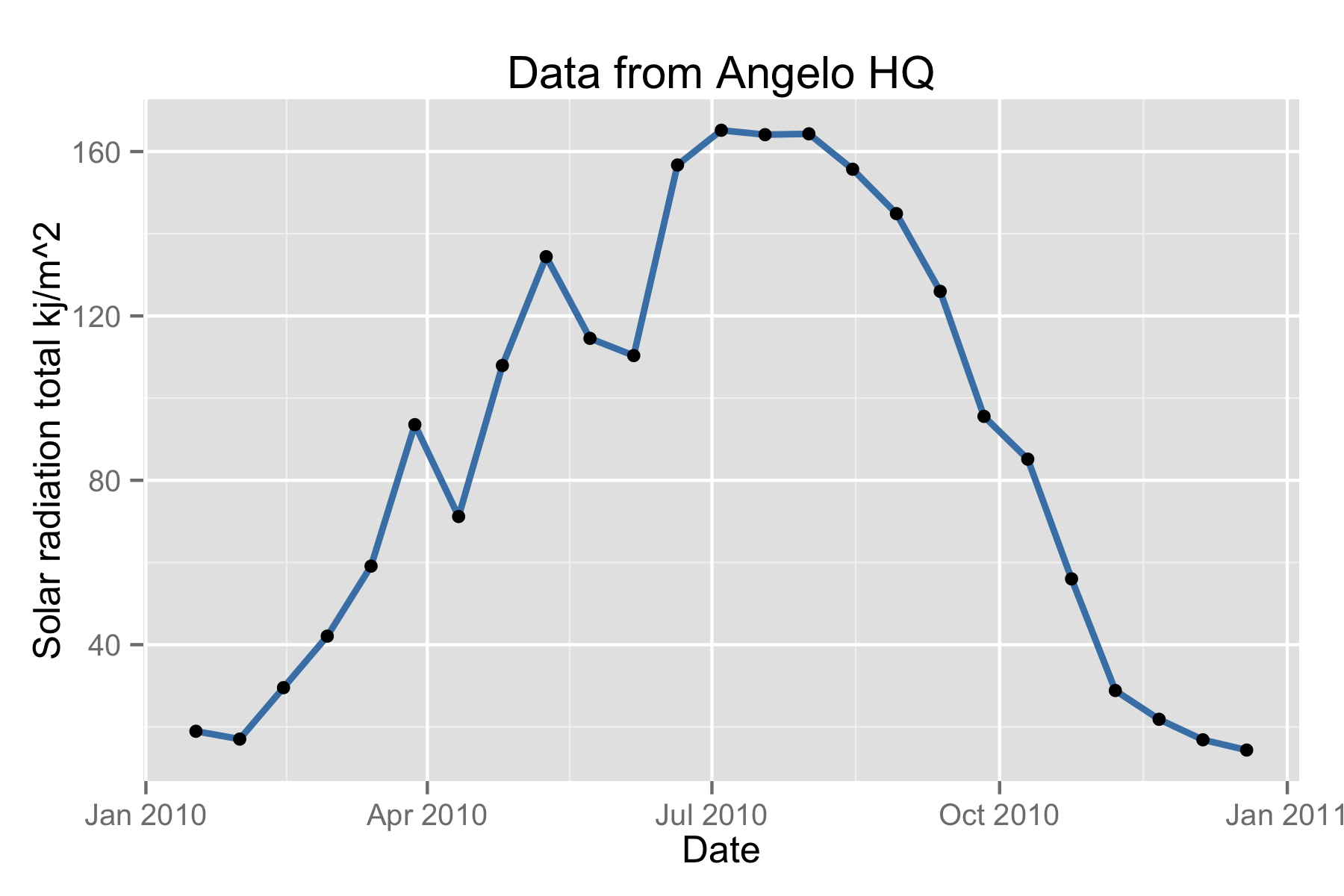
head(ee_list_sensors())
## station_name units ## 1 Angelo HQ WS Kilojoules per square meter ## 2 Angelo Meadow WS Watts per square meter ## 3 Angelo HQ SF Eel Gage Percent ## 4 Angelo HQ WS Degree ## 5 Cahto Peak WS Meters per second ## 6 Angelo Meadow WS Meters per second ## variable method_name record ## 1 Solar radiation total kj/m^2 Conversion to 30-minute timesteps 1625 ## 2 Solar radiation total w/m^2 Conversion to 30-minute timesteps 1632 ## 3 Rel humidity perc Conversion to 30-minute timesteps 1641 ## 4 Wind direction degrees Conversion to 30-minute timesteps 1644 ## 5 Wind speed avg ms Conversion to 30-minute timesteps 1651 ## 6 Wind speed max ms Conversion to 30-minute timesteps 1654
# First we can grab the list of sensor ids sensor_ids <- ee_list_sensors()$record # In this case we just need data for sensor with id 1625 angelo_hq <- sensor_ids[1] results <- ee_sensor_data(angelo_hq, page = 2, progress = FALSE) results
## [Total results]: 56779 ## [Args]: ## page_size = 25 ## page = 2 ## [Type]: sensor ## [Number of results]: 25
stations <- ee_list_sensors() sensor_df <- ee_sensor_agg(sensor_id = stations[1, c("record")], weeks = 2, progress = FALSE) head(sensor_df$data)
## begin_date mean min max sum count ## 2 2010-01-17 18.94 0 150.8 7613 402 ## 26 2010-01-31 17.03 0 237.7 11444 672 ## 3 2010-02-14 29.54 0 336.3 19852 672 ## 4 2010-02-28 42.08 0 402.5 28276 672 ## 5 2010-03-14 59.12 0 466.6 39730 672 ## 6 2010-03-28 93.55 0 490.6 62678 670
rodent_pages <- ee_pages(ee_observations(order = "Rodentia", progress = FALSE, quiet = TRUE)) page_breaks <- split(1:rodent_pages, ceiling(seq_along(1:rodent_pages)/1000)) rodent_data <- list() # This step will take a while, as it downloads ~50 mb of data with pauses to # avoid hammering API for (i in 1:length(rodent_data)) { rodent_data <- ee_observations(order = "Rodentia", page = page_breaks[[1]]) write.csv(rodent_data$data, file = paste0("rodent_data", i, ".csv")) }
library(plyr) rodent_pages <- ee_pages(ee_observations(order = "Rodentia", progress = FALSE, quiet = TRUE)) page_breaks <- split(1:rodent_pages, ceiling(seq_along(1:rodent_pages)/1000)) results <- ldply(page_breaks, function(x) { results <- ee_pages(ee_observations(order = "Rodentia", page = x, quiet = TRUE)) results$data }, .progress = "text")
library(dplyr) rodents_summary <- rodents %.% filter(!is.na(year)) %.% group_by(state = abbreviation, year) %.% summarise(counts = n()) head(rodents_summary)
## State Year Counts ## 1 CA 1818 9 ## 2 OR 1899 16 ## 3 CA 1904 178 ## 4 OR 1905 32 ## 5 CA 1905 18 ## 6 CA 1907 32
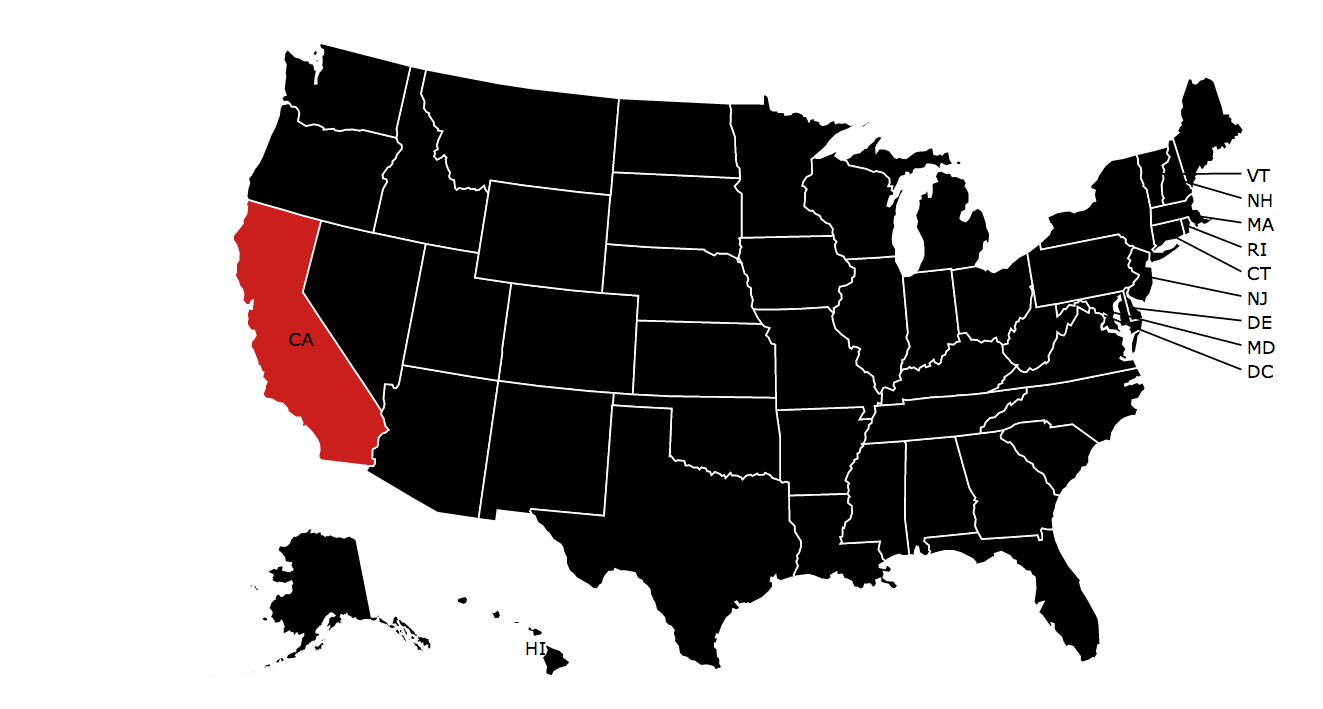
library(rMaps) rodents_summary <- dplyr::filter(rodents_summary, Year > 1899) ichoropleth(Counts ~ State, data = rodents_summary, animate = "Year", pal = "Reds", ncuts = 4, play = TRUE)
all_lists <- ee_checklists() nrow(all_lists)
## [1] 57
unique(all_lists$subject)
## [1] "Mammals" "Mosses" ## [3] "Beetles" "Spiders" ## [5] "Amphibians" "Ants" ## [7] "Fungi" "Lichen" ## [9] "Plants" "Reptiles" ## [11] "Algae" "Arthropods" ## [13] "Liverworts" "Birds" ## [15] "Butterflies & Moths" "Fish" ## [17] "Bees" "Archaeognatha" ## [19] "Insects 1937-1955" "Orthoptera and Cicadidae" ## [21] "Butterflies" "Aquatic Insects" ## [23] "Stoneflies" "Caddisflies" ## [25] "whipworms" "Tardigrades" ## [27] "Small Mammals" "False Scorpions" ## [29] "Crane-flies" "Chiggers" ## [31] "Cestodes" "Rodents" ## [33] "Lichens" "Grasses and Forbes" ## [35] "Macroinvertebrates" "Amphibians and Reptiles"
spiders <- ee_checklists(subject = "Spiders") spiders
## record ## 4 bigcb:specieslist:15 ## 10 bigcb:specieslist:20 ## footprint ## 4 http://ecoengine.berkeley.edu/api/footprints/hastings-reserve/ ## 10 http://ecoengine.berkeley.edu/api/footprints/angelo-reserve/ ## url ## 4 http://ecoengine.berkeley.edu/api/checklists/bigcb%3Aspecieslist%3A15/ ## 10 http://ecoengine.berkeley.edu/api/checklists/bigcb%3Aspecieslist%3A20/ ## source subject ## 4 http://ecoengine.berkeley.edu/api/sources/18/ Spiders ## 10 http://ecoengine.berkeley.edu/api/sources/18/ Spiders
library(plyr) spider_details <- ldply(spiders$url, checklist_details) names(spider_details)
## [1] "url" "observation_type" ## [3] "scientific_name" "collection_code" ## [5] "institution_code" "country" ## [7] "state_province" "county" ## [9] "locality" "coordinate_uncertainty_in_meters" ## [11] "begin_date" "end_date" ## [13] "kingdom" "phylum" ## [15] "clss" "order" ## [17] "family" "genus" ## [19] "specific_epithet" "infraspecific_epithet" ## [21] "source" "remote_resource" ## [23] "earliest_period_or_lowest_system" "latest_period_or_highest_system"
photos <- ee_photos(quiet = TRUE, progress = FALSE) photos
## [Total results]: 43708 ## [Args]: ## page_size = 25 ## georeferenced = 0 ## page = 1 ## [Type]: photos ## [Number of results]: 25
view_photos()
poppys <- ee_photos(scientific_name = "Eschscholzia californica", quiet = TRUE, progress = FALSE) view_photos(poppys)
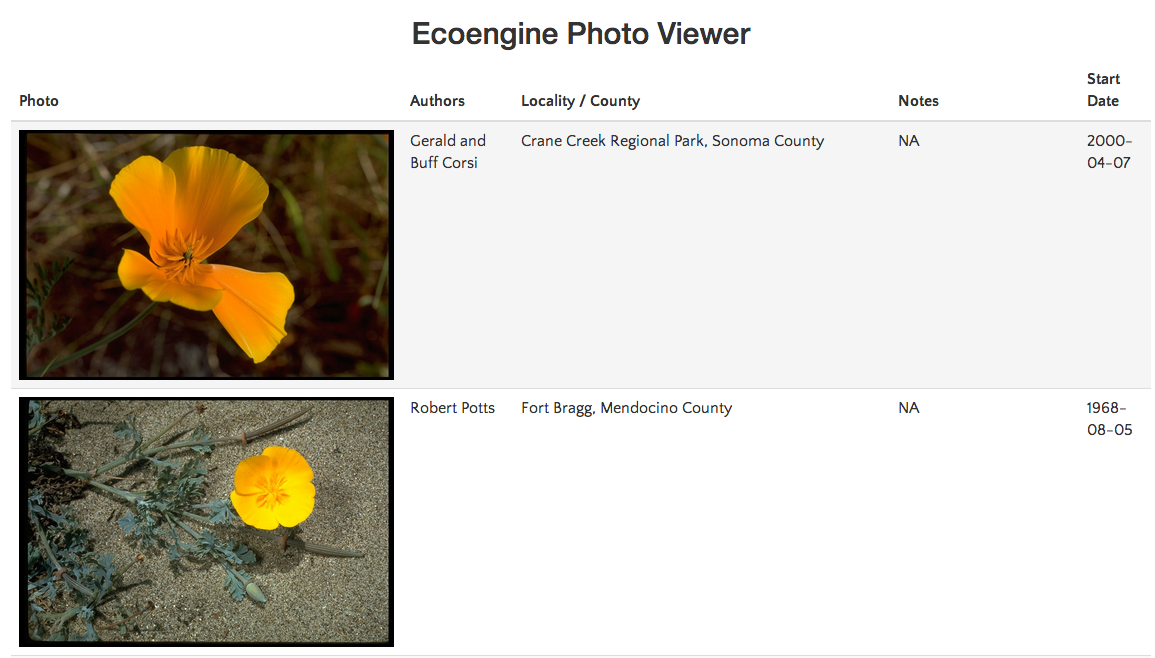
/
#